Gausian Process Regression with Covariance Product and Sum#
The atmospheric CO2 concentration readings in parts per million (ppm) by volume from air samples are collected continuously by the Mauno Loa observatory in Hawaii. The data are freely available at ftp: // ftp.cmdl.noaa.gov/ccg/co2/trends/. http://scrippsco2.ucsd.edu/data/atmospheric_co2/mlo
This example was first presented in the book of Rasmussen & Williams [1] and serves as a tutorial for kernel composition, see for instance - https://docs.pymc.io/notebooks/GP-MaunaLoa.html - https://scikit-learn.org/stable/auto_examples/gaussian_process/plot_gpr_co2.html - http://dfm.io/george/current/user/hyper/
The modelling and the SDE representation of Arno Solin and Simo Särkkä [2], which differs from the book of Rasmussen & Williams [3], is used here.
[1]:
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from pysip.regressors import Regressor
from pysip.statespace import Matern32, Matern52, Periodic
Load and prepare the Mauna Loa CO2 data set#
[2]:
df = pd.read_csv(
"../../../data/mauna_loa/monthly_in_situ_co2_mlo.csv",
comment='"',
header=0,
skipinitialspace=True,
)
df = df[["Date.1", "CO2"]].set_index("Date.1")
df.index.name = "Date"
df = df[df.CO2 != -99.99]
# Use data until 2010 for the fit and the rest for the prediction
C02_mean = df["CO2"].mean()
df["CO2_fit"] = df["CO2"] - C02_mean
df["CO2_pred"] = df["CO2_fit"]
df["CO2_pred"][df.index >= 2010] = np.nan
Initialize parameters and models#
[3]:
# Matérn 5/2 for slow rising trend (long-term effects)
p1 = [
dict(name="mscale", value=2.295e03, transform="log"),
dict(name="lscale", value=4.864e02, transform="log"),
dict(name="sigv", value=2.210e-01, transform="log"),
]
# Periodic * Matérn 3/2 for quasi-periodic variations
p2 = [
dict(name="period", value=1.0, transform="fixed"),
dict(name="mscale", value=2.833e00, transform="log"),
dict(name="lscale", value=1.341e00, transform="log"),
dict(name="sigv", value=0.0, transform="fixed"),
]
p3 = [
dict(name="mscale", value=1.0, transform="fixed"),
dict(name="lscale", value=2.607e02, transform="log"),
dict(name="sigv", value=0.0, transform="fixed"),
]
# Matérn 3/2 for short-term effects
p4 = [
dict(name="mscale", value=4.595e-01, transform="log"),
dict(name="lscale", value=6.359e-01, transform="log"),
dict(name="sigv", value=0.0, transform="fixed"),
]
k1 = Matern52(p1, name="k1")
k2 = Periodic(p2, name="k2")
k3 = Matern32(p3, name="k3")
k4 = Matern32(p4, name="k4")
# Compose covariance function
K = k1 + k2 * k3 + k4
K
[3]:
GPSum(hold_order=0, method='mfd', name='k1__+__k2__x__k3__+__k4')
Initialize the regressor#
[4]:
reg = Regressor(K, outputs="CO2_fit")
Do the frequentist fit#
[5]:
fit_summary, corr_matrix, opt_summary = reg.fit(df=df)
fit_summary
[ ]:
tnew = np.arange(1958, 2030, 0.01)
ds = reg.predict(df=df, smooth=True, tnew=tnew)
[ ]:
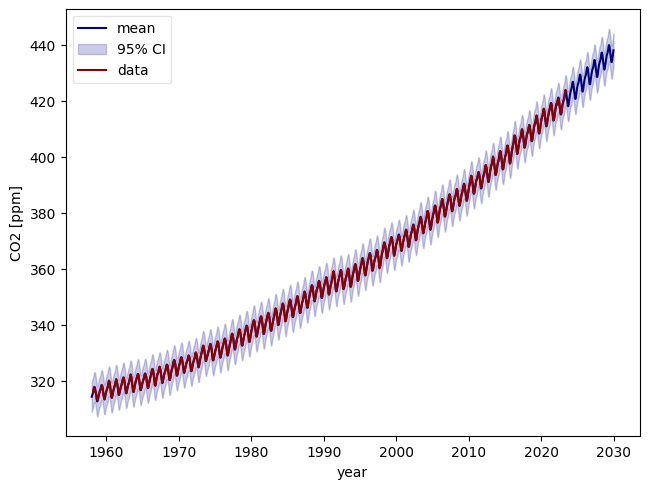
# Plot output mean and 95% credible intervals
ym = ds["y_mean"].sel(outputs="CO2_fit")
ysd = ds["y_std"].sel(outputs="CO2_fit")
plt.plot(tnew, C02_mean + ym, color="navy", label="mean")
plt.fill_between(
tnew,
C02_mean + ym - 2 * ysd,
C02_mean + ym + 2 * ysd,
color="darkblue",
alpha=0.2,
label=r"95% CI",
)
plt.plot(df.index, df["CO2"], color="darkred", label="data")
plt.tight_layout()
plt.legend(loc="best", fancybox=True, framealpha=0.5)
ax = plt.gca()
ax.set_xlabel("year")
ax.set_ylabel("CO2 [ppm]")
Text(38.347222222222214, 0.5, 'CO2 [ppm]')